Regulation of pathogenic and sexual development in Ustilago maydis
The basidiomycete Ustilago maydis is one of the prime model systems to study plant/pathogen interactions. Similar to other pathogens of plants and humans, U. maydis is a dimorphic fungus, with haploid yeast like cells, called sporidia, that grow strictly saprophytic, and a filamentous dikaryon that is dependent on the host plant corn for its development. After plant infection, U. maydis induces the formation of tumors in which the fungal hyphae differentiate and generate diploid spores. The outstanding feature of the U. maydis system is that the change from saprophytic growth to the biotrophic stage is controlled by a single genetic switch, namely the b-mating type locus encoding the two transcription factors bE and bW.
The major interest of our group is the elucidation of the regulatory processes that lead to the establishment of the pathogenic stage. Our main aim is the identification and characterization of genes controlled by the bE/bW transcription factor. We presume that these genes play a crucial role in the morphological switch as well as in pathogenic development.
To get a comprehensive view of b-dependent gene regulation and regulatory networks, we have established DNA arrays for U. maydis that allow to monitor the expression of about 6200 U. maydis genes in parallel. We have employed the gene chip technology to monitor the changes in gene expression after formation of an active bE/bW heterodimer in a 12 hours time course, and have identified more than 300 b-responsive genes. The bioinformatic analysis of these genes allowed to visualize the different processes controlled by the b-locus. A large number of b-regulated genes affect cell cycle, mitosis and DNA replication, which is consistent with the observation that cell division is stalled after b-induction, until the fungus has entered the plant. Other genes are predicted to be involved in the synthesis and modification of cell wall components, making it likely that the cell wall of the yeast-like sporidia and of the filamentous dikaryon are dissimilar in their overall composition.
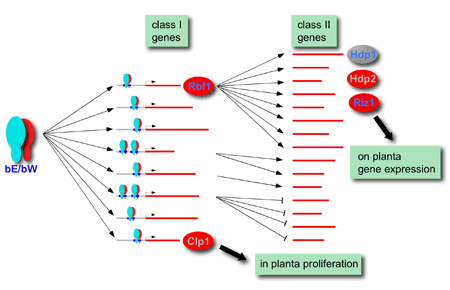
b-dependent regulatory network. Class I genes are directly regulated by bE/bW via binding to conserved b-binding sequences (bbs) within the promoter regions. The majority of the indirectly regulated class II genes are controlled by the b-dependent transcription factor Rbf1. Clp1 is expressed independently from Rbf1 and required for in planta proliferation. The expression of the transcription factors Hdp1, Hdp2 and Riz1 is dependent on Rbf1. Riz1 is necessary for the induction of various genes on the plant surface. Red indicates that the proteins are obligatory for pathogenic development, blue letters specify that induced expression leads to a cell cycle arrest.
Among the b-regulated genes we have focused so far on genes with potential regulatory functions. This has led to the identification of four novel pathogenicity genes: riz1, rbf1 and hdp2, encoding two zinc finger and one homeodomain transcription factors, respectively, and clp1, encoding a protein with unknown function. Rbf1 is required for both filamentous growth as well as pathogenic development. Microarray experiments revealed that this transcription factor plays a crucial role for the expression of a large fraction of the b-regulated genes, and thus has a central role within the b-regulatory network. riz1, one of the Rbf1 dependent genes, is required for the expression of genes specifically induced on the plant surface. A large fraction of the riz1 dependent genes code for U. maydis specific proteins that are predicted to be secreted. We hypothesize that these proteins may be involved in suppressing plant defense reactions and reprogramming of the host. clp1 might be required for the release of the cell cycle arrest in planta, since clp1 mutant strains are able to infect, but are blocked in development before the first cell division. We are currently evaluating the crosstalk between the different transcription factors and aim to identify the specific input/output signals and their connection to the establishment of the pathogenic program.
Another focus in our research has been to understand the impact of chromatin mediated transcriptional regulation on pathogenic development. We have previously identified the histone deacetylase Hda1 and Rum1, a protein with similarities to the human retinoblastoma binding protein 2, that are both required for the formation of teliospores. Data from expression profiles and co-immunoprecipitation experiments place Hda1 and Rum1 in one complex. The complex is required for the repression of a significant number of genes that are induced in tumor tissue in wild type cells. Interestingly, several of these genes are clustered, emphasizing that the regulation may occurs at the level of chromatin structure.
Projects
The following projects are currently under investigation:
Regulating early infection and in planta development of Ustilago maydis
Modulation of transcription factors by Clp1 specifies mating-type dependent signaling during biotrophic development of Ustilago maydis

